Connections to Transcription Factors and a Massive Setback
This has been a week of progress and setbacks. I was trying to create connections between known transcription factors and the genes in my dataset.
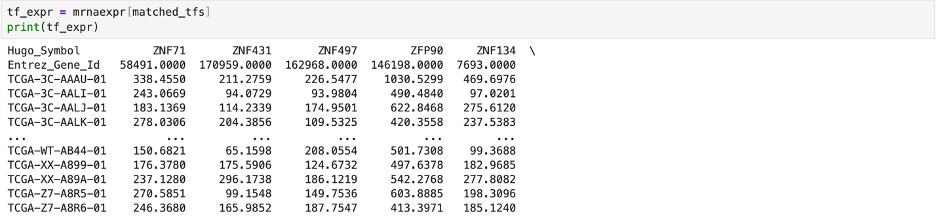
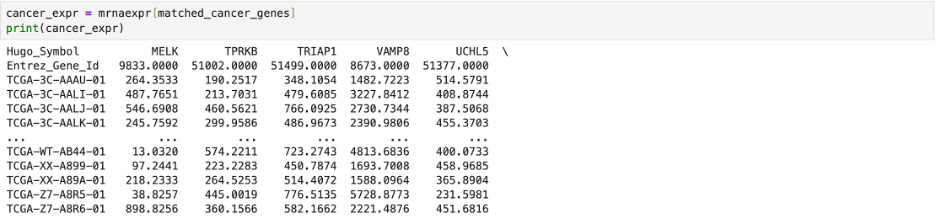
First, I downloaded data of known cancer genes and transcription factors. I then used the DataFrame.columns function in pandas to create a list including the Hugo Symbols (identifiers) for genes found in the experimental breast cancer dataset.
The next step was to find matches between the experimental data and the known data. Using the intersection function, I found the intersections between the experimental data set with the cancer genes and transcription factors respectively. In doing this, I found 119 matches out of 249 cancer genes and 1020 matches out of 1640 transcription factors.
Then, I used the matching genes to splice the original transcription factor data and save it under new lists. One list (tf_expr in the first code snippet below) was the experimental expression of the matching known transcription factors while the other list (cancer_expr in the second code snipped below) was the experimental expression of known cancer related genes.
I then used the Lasso model in the scikit library to find correlations between transcription factors. The Lasso model is a statistical model that analyzes data to identify important values and predicts an outcome. In this case, the model predicted expression of a certain gene and which transcription factors were the most connected to the gene’s expression. Essentially, I created a basic model to predict networks of transcription factors. For example, I mapped 30 genes that were predicted to be connected to TP53: And TP53 is just one example. I was also able to map other genes such as MYC and VDR.
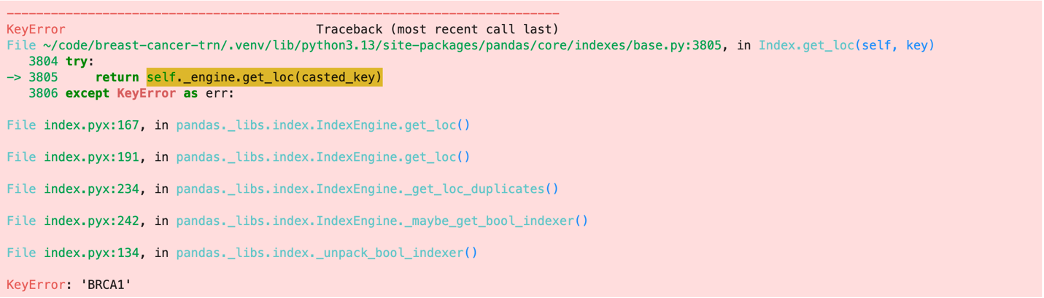
This is where the trouble started, though. After mapping those three genes, I tried mapping BRCA1. That is when I realized that my experimental data set did not have BRCA1 data. This was not the only missing gene. Other breast cancer related genes such as FOXA1, ESR1, GATA3, and PALB2, to name a few, were also missing. This is when I realized that the experimental dataset that I have been using has been filtered of some key protein coding regions. This means that I am missing out on some important genetic networks.
I have found a more comprehensive genetic network, though, so next week I will redo the work I have already done with the new data. This should hopefully be done faster since I have already done it once and my school workload is reducing as the end of the school year approaches.
After I am done redoing the progress I have made, I will use Lasso to analyze all known matching cancer genes in the list. Then I will compare the transcription factor networks I find to known networks. This will tell me whether what I have found is known or new.
Thank you so much for reading and please stay tuned for future updates!